Project Proposal
Model Proposal for Life from Scratch
Bhaskar Kumawat
- Course ID: CMPLXSYS 530
- Course Title: Computer Modeling of Complex systems
- Term: Winter, 2025
Overview
Goal
I wish to model the origin of self-replication in a 2D world consisting of atoms that move and bond with each other based on some physical and chemical rules. My goal for this class project would be to get any sort of self-replication in this system after millions of time-steps, but I wish to extend it to get Replication Involving Replicable Imperfections (RIRI). RIRI is characterised by molecules (i.e. groups of bonded atoms) creating robust copies while also copying any errors that may have appeared previously in the process (Benner 2014).
Rationale
Out of all possible chemistries (set of bonding rules between species of atoms) that can exist, only some are likely to give rise to self-replicating molecules. Further, the environmental conditions (other atoms in the surroundings, temperature etc.) can vitally affect whether a chemistry is able to sustain self-replication. This project aims to explore the conditions that are required for self-replication to arise in a simple physical system, given arbitrary interaction rules and environments. I believe that a 2D world consisting of atoms that follow some physics-y rules (eg. diffusion and collisions) should be sufficient to observe these phenomena. Indeed, simpler, yet similar grid-based simulations have shown this “origin of self-replication” for one particular set of rules (Hutton 2002). However, I wish to test this for a large number of randomly generated chemistries to test some general hypothesis about origin of life.
Main Micro-level Processes and Macro-level Dynamics of Interest
The simulation world that I’m building consists of a 2D plane over which a large number of circular “atom” agents reside. Each atom has a species and a state (both being integer values between 0 and 255). The main processes that these atoms undergo and the expected emergent dynamics are as follows:
Micro-level processes
- Atomic Diffusion: Each atom performs a random 2D motion similar to a Brownian particle in a viscous fluid.
- Atomic Collisions: Atoms are solid bodies and collide with each other when they come in contact.
- Bonding & bond-breaking: The program allows the user to specify any chemistry in terms of creation and decomposition of bonds between atoms. Bonds are created and broken probabilistically based on collisions. If two atoms of given species and state (as specified by the chemistry) collide, they can form a bond with some probability. On the other hand, if two atoms of given species and state (also specified in the chemistry) are already bonded, they may break with some probability.
Macro-level/emergent dynamics
- Self-replication: Self-replication is described as a process where groups of atoms (or molecules) are able to sustain continual creation of self-similar copies in the simulation world. Previous work has shown this is indeed possible in a similar system, but only for a very specific set of bonding rules (Hutton 2002).
- Evolution: Evolution is the process by which these molecules change as they self-replicate, but in a way that these errors are retained in future replications. Thus, RIRI (Replication involving replicable imperfections) is an important requirement for us to observe evolution in this system (Benner 2014; Fontana and Schuster 1998).
Model
The model consists of a 2D plane with circular atoms that move around and collide with each other. Collisions can lead to “reactions” where two atoms (assuming they are compatible) can bond with each other to form a molecule. Molecules can also decompose into atoms based on the reactions that are allowed. The set of reactions being used for a given simulation is called a “chemistry”. The following diagram shows an example chemistry and the resulting simulation world at a particular time.
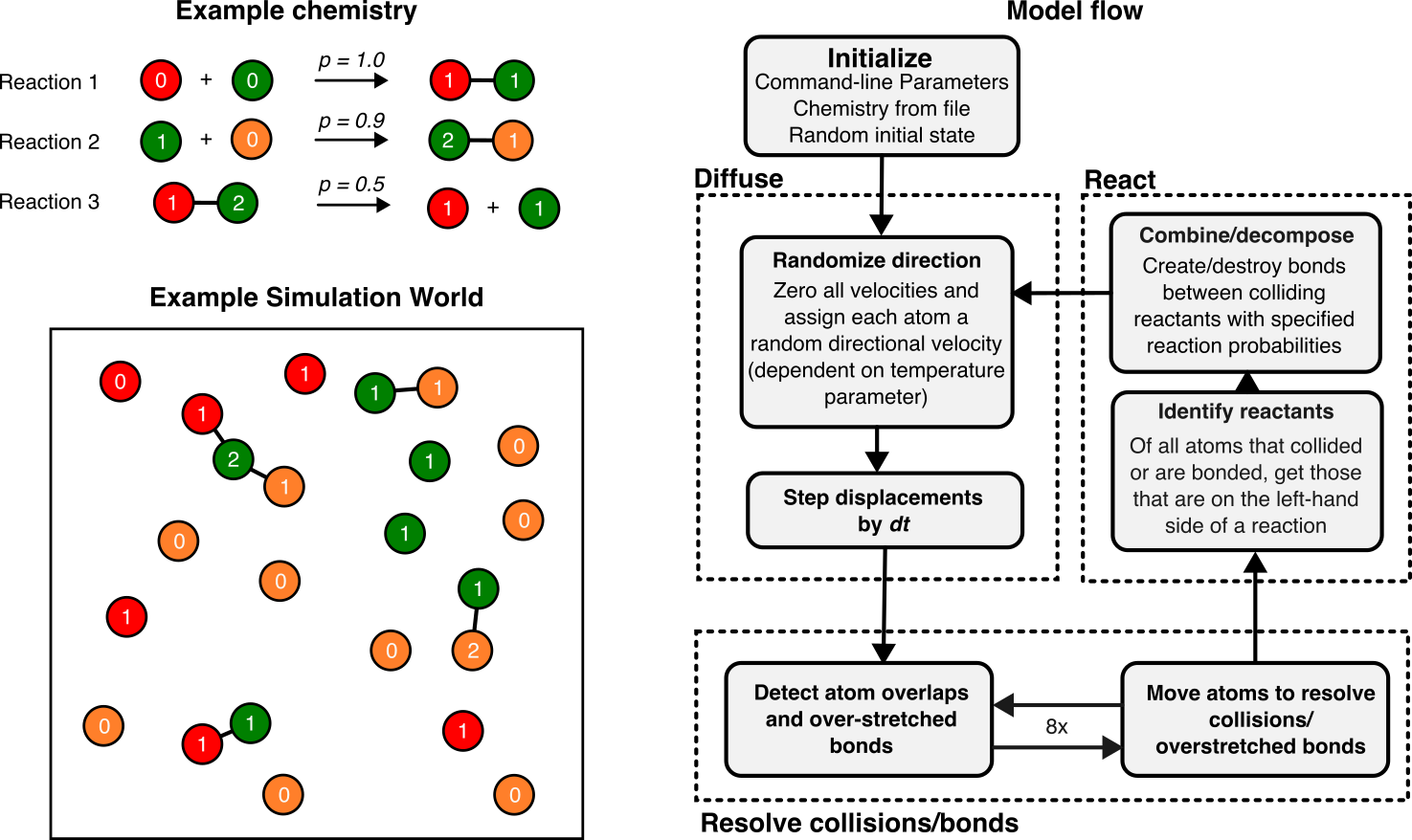
1) Environment
The will have a finite, continuous 2D space with reflecting boundaries (i.e. walls). For now, I’m keeping the temperature constant across the world, but I envision having the temperature as a continuous function over the simulation environment. I also plan to add some sources and sinks in the world for atoms to enter and leave the system, thus allowing some sort of “selection” to emerge naturally.
2) Agents
The agents are circular “atoms” in a 2D world. All agents are the same size and have both a position in the world and a velocity (both of which are 2-dimensional float vectors). Each atom also has a “species” and a “state” property, both of which are integral values between 0 and 255. The species of an atom remains constant throughout the simulation but the state can change during reactions. Atoms diffuse through the world performing a random walk and can react with other atoms when they collide. These reactions can either form a “bond” between atoms, joining them together as they move around the world, or simply change their state without bonding. Bonded atoms (a molecule) can also decompose into their constituents. Interactions are entirely dependent on collisions and thus on spatial proximity between the atoms.
3) Model Scheduling
A schematic of the model schedule is shown in figure Figure 1. The entire setup can be essentially divided into four parts.
- Initialization: Here, the program initializes the parameters and chemistries required for the simulation. It also generates a random initial state for the system based on a random number seed and the initial number of atoms (provided as parameters).
The following steps are repeated until the user ends the simulation:
- Diffusion: Here, each atom is assigned a random velocity in any direction (with magnitude proportional the the
temperatureparameter). Then, the simulation is progressed by a single time-step (of length \(dt\)) so the velocity change appears as a displacement of the atoms. - Resolve collisions/bonds: In this step, the simulation takes note of all collisions (overlapping atoms) and bond extensions and tries to resolve them by moving the atoms by some computed distance. This step is performed 8x times because sometimes resolving collision between two atoms may create other collisions in the world.
- Reaction: Here, all atom pairs that collided with each other are checked for a “reaction” by looking their species/states up in the user specified chemistry. If two atoms can react, they are either bonded or their state is changed probabilistically based on the reaction probability (also specified by the user). Bonded atoms are also checked for decomposition.
4) Model Parameters and Initialization
The main user defined parameters (apart from the chemistry) are as follows:
size_x,size_y: Size of the simulation world in arbitrary units.diameter: Diameter of an atomtemperature: Temperature of the world, determines how random an atoms’ movement is.init_atoms: Initial number of atoms in the world.
The model is initialized by first obtaining these parameters from the user through the command line. Then, the program randomly samples init_atoms number of locations in the simulation world to create the initial world state.
5) Assessment and Outcome Measures
For the purpose of quantifying self-replication, I will output all molecules and their numbers that are present in the system every few time-steps. These numbers can be simply plotted over time to check if there is a sudden explosive increase in the population of a particular molecule, indicating self-replication.
I will perform a similar analysis with a lot of randomly generated chemistries to find chemistries that allow self-replication. Then, I will narrow down on chemistries where the succesion between replicating molecules is such that it qualifies for the definition of replication with replicable imperfections: i.e., new self-replicators are similar to old self-replicators but are also stable and do not revert to an earlier state.
6) Parameter Exploration
The most interesting parameter to vary here would be temperature, as it allows an increase in the possiblity of chance encounters between far apart molecules and atoms. However, instead of varying the parameter between simulations, I might opt for a larger world with heterogeneous temperature at different points in the world. I hypothesis that a more “hetergeneous” world such as this would lead to self-replication faster than a homogeneous temperature simulation.
Questions and challenges
Question
- Rigid vs flexible bonds: There are essentially two ways to create a bond between atoms. In the first case, the atoms are allowed to rotate around the bond and a large molecule can essentially flop around in the world and does not have a rigid 2D structure. On the other hand, a rigid bond restricts the rotation of the atoms and fixes the structure of the molecule to what it was when the bond was formed. I feel like the flexible case would be more interesting (both visually and in terms of allowing self-replication) but I’m not sure if that’s realistic (because real molecules are actually somewhat rigidly bonded to each other).
Challenge
- Moving to a larger simulation: According to my tests, the simulation can support around 100,000 atoms at decent speed on a somewhat powerful personal computer. I would like to scale this further and go upto maybe 1-10 million particles. I’m not sure about the techniques I could use for this, maybe performing the simulation on a GPU?
Code
The code is available in the github repository here (branch bevylife). The code is written in Rust and uses the Bevy game engine and the Rapier physics library for improved performance (I tried writing these from scratch first but it’s both time consuming and not as performative as using a pre-built engine). I have implemented atomic diffusion and collisions as of now. The collisions are also reported as events that I can use to make bonds. Rigid bonding (where the bonds are NOT “floppy”) was easy to implement but is not as interesting. I’m currently trying to implement “floppy” bonds (a revolute joint). Pre-built binaries to run the current simulation on different platforms are available here:
- Linux: 64-bit
- Windows: 64-bit
- MacOS: M-series Processors (eg. M1-M4), Intel
If you’re not sure which one to get, just choose the first link for your operating system. If that doesn’t work, you can try the next one. Please have a look at the release page here for instructions on running the program.